This is a concordance module that will assess consistency within a patient
record by evaluating the presence or absence of two user-provided clinical
events (prc_event_file). The checks in the module will establish whether
one of, neither, or both events are present in each patient record and
summarize these results across the full cohort. A sample version of the
input file is accessible with patientrecordconsistency::. This function
is compatible with both the OMOP and the PCORnet CDMs based on the user's
selection.
Usage
prc_process(
cohort,
prc_event_file,
omop_or_pcornet,
multi_or_single_site = "single",
anomaly_or_exploratory = "exploratory",
age_groups = NULL,
patient_level_tbl = FALSE,
fu_breaks = c(0, 1, 3, 8, 11, 15, 25, 50, 100),
p_value = 0.9,
time = FALSE,
time_span = c("2012-01-01", "2020-01-01"),
time_period = "year"
)Arguments
- cohort
tabular input || required
The cohort to be used for data quality testing. This table should contain, at minimum:
site| character | the name(s) of institutions included in your cohortperson_id/patid| integer / character | the patient identifierstart_date| date | the start of the cohort periodend_date| date | the end of the cohort period
Note that the start and end dates included in this table will be used to limit the search window for the analyses in this module.
- prc_event_file
tabular input || required
A table with the definitions of each of the events. This table should contain two rows, one for each event, with the following columns:
event| character | a string, either A or B, representing the event type. A will be treated as the event that is expected to occur first in a sequenceevent_label| character | a descriptive label for the eventdomain_tbl| character | the name of the CDM table where the event is definedconcept_field| character | the string name of the field in the domain table where the concepts are locateddate_field| character | the name of the field in the domain table with the date that should be used for temporal filteringvocabulary_field| character | for PCORnet applications, the name of the field in the domain table with a vocabulary identifier to differentiate concepts from one another (ex: dx_type); can be set to NA for OMOP applicationscodeset_name| character | the name of the codeset that defines the event of interestfilter_logic| character | logic to be applied to the domain_tbl in order to achieve the definition of interest; should be written as if you were applying it in a dplyr::filter command in R
To see an example of this input, see
?patientrecordconsistency::prc_event_file- omop_or_pcornet
string || required
A string, either
omoporpcornet, indicating the CDM format of the dataomop: run theprc_process_omop()function against an OMOP CDM instancepcornet: run theprc_process_pcornet()function against a PCORnet CDM instance
- multi_or_single_site
string || defaults to
singleA string, either
singleormulti, indicating whether a single-site or multi-site analysis should be executed- anomaly_or_exploratory
string || defaults to
exploratoryA string, either
anomalyorexploratory, indicating what type of results should be produced.Exploratory analyses give a high level summary of the data to examine the fact representation within the cohort. Anomaly detection analyses are specialized to identify outliers within the cohort.
- age_groups
tabular input || defaults to
NULLIf you would like to stratify the results by age group, create a table or CSV file with the following columns and use it as input to this parameter:
min_age| integer | the minimum age for the group (i.e. 10)max_age| integer | the maximum age for the group (i.e. 20)group| character | a string label for the group (i.e. 10-20, Young Adult, etc.)
If you would not like to stratify by age group, leave as
NULL- patient_level_tbl
boolean || defaults to
FALSEA boolean indicating whether an additional table with patient level results should be output.
If
TRUE, the output of this function will be a list containing both the summary and patient level output. Otherwise, this function will just output the summary dataframe- fu_breaks
vector || defaults to
c(0, 1, 3, 8, 11, 15, 25, 50, 100)A numeric vector that defines how to group different windows of follow up time. This parameter is used for both
Anomaly Detection, Cross-Sectionalchecks- p_value
numeric || defaults to
0.9The p value to be used as a threshold in the Multi-Site, Anomaly Detection, Cross-Sectional analysis
- time
boolean || defaults to
FALSEA boolean to indicate whether to execute a longitudinal analysis
- time_span
vector - length 2 || defaults to
c('2012-01-01', '2020-01-01')A vector indicating the lower and upper bounds of the time series for longitudinal analyses
- time_period
string || defaults to
yearA string indicating the distance between dates within the specified time_span. Defaults to
year, but other time periods such asmonthorweekare also acceptable
Value
This function will return a dataframe summarizing the co-occurrence of events within a patient record. For a more detailed description of output specific to each check type, see the PEDSpace metadata repository
Examples
#' Source setup file
source(system.file('setup.R', package = 'patientrecordconsistency'))
#' Create in-memory RSQLite database using data in extdata directory
conn <- mk_testdb_omop()
#' Establish connection to database and generate internal configurations
initialize_dq_session(session_name = 'prc_process_test',
working_directory = my_directory,
db_conn = conn,
is_json = FALSE,
file_subdirectory = my_file_folder,
cdm_schema = NA)
#> Connected to: :memory:@NA
#' Build mock study cohort
cohort <- cdm_tbl('person') %>% dplyr::distinct(person_id) %>%
dplyr::mutate(start_date = as.Date(-5000), # RSQLite does not store date objects,
# hence the numerics
end_date = as.Date(15000),
site = ifelse(person_id %in% c(1:6), 'synth1', 'synth2'))
#' Build function input table
prc_events <- tidyr::tibble(event = c('a', 'b'),
event_label = c('hypertension', 'inpatient/ED visit'),
domain_tbl = c('condition_occurrence', 'visit_occurrence'),
concept_field = c('condition_concept_id', 'visit_concept_id'),
date_field = c('condition_start_date', 'visit_start_date'),
vocabulary_field = c(NA, NA),
codeset_name = c('dx_hypertension', 'visit_edip'),
filter_logic = c(NA, NA))
#' Execute `prc_process` function
#' This example will use the single site, exploratory, cross sectional
#' configuration
prc_process_example <- prc_process(cohort = cohort,
multi_or_single_site = 'single',
anomaly_or_exploratory = 'exploratory',
time = FALSE,
omop_or_pcornet = 'omop',
prc_event_file = prc_events) %>%
suppressMessages()
#> ┌ Output Function Details ──────────────────────────────────────┐
#> │ You can optionally use this dataframe in the accompanying │
#> │ `prc_output` function. Here are the parameters you will need: │
#> │ │
#> │ Always Required: process_output │
#> │ │
#> │ See ?prc_output for more details. │
#> └───────────────────────────────────────────────────────────────┘
prc_process_example
#> # A tibble: 5 × 8
#> site event_a_num event_a_name event_b_num event_b_name pt_ct total_pts
#> <chr> <dbl> <chr> <dbl> <chr> <int> <int>
#> 1 combined 0 hypertension 0 inpatient/ED vi… 6 12
#> 2 combined 0 hypertension 1 inpatient/ED vi… 1 12
#> 3 combined 1 hypertension 1 inpatient/ED vi… 2 12
#> 4 combined 1 hypertension 2 inpatient/ED vi… 2 12
#> 5 combined 1 hypertension 5 inpatient/ED vi… 1 12
#> # ℹ 1 more variable: output_function <chr>
#' Execute `prc_output` function
prc_output_example <- prc_output(process_output = prc_process_example)
prc_output_example
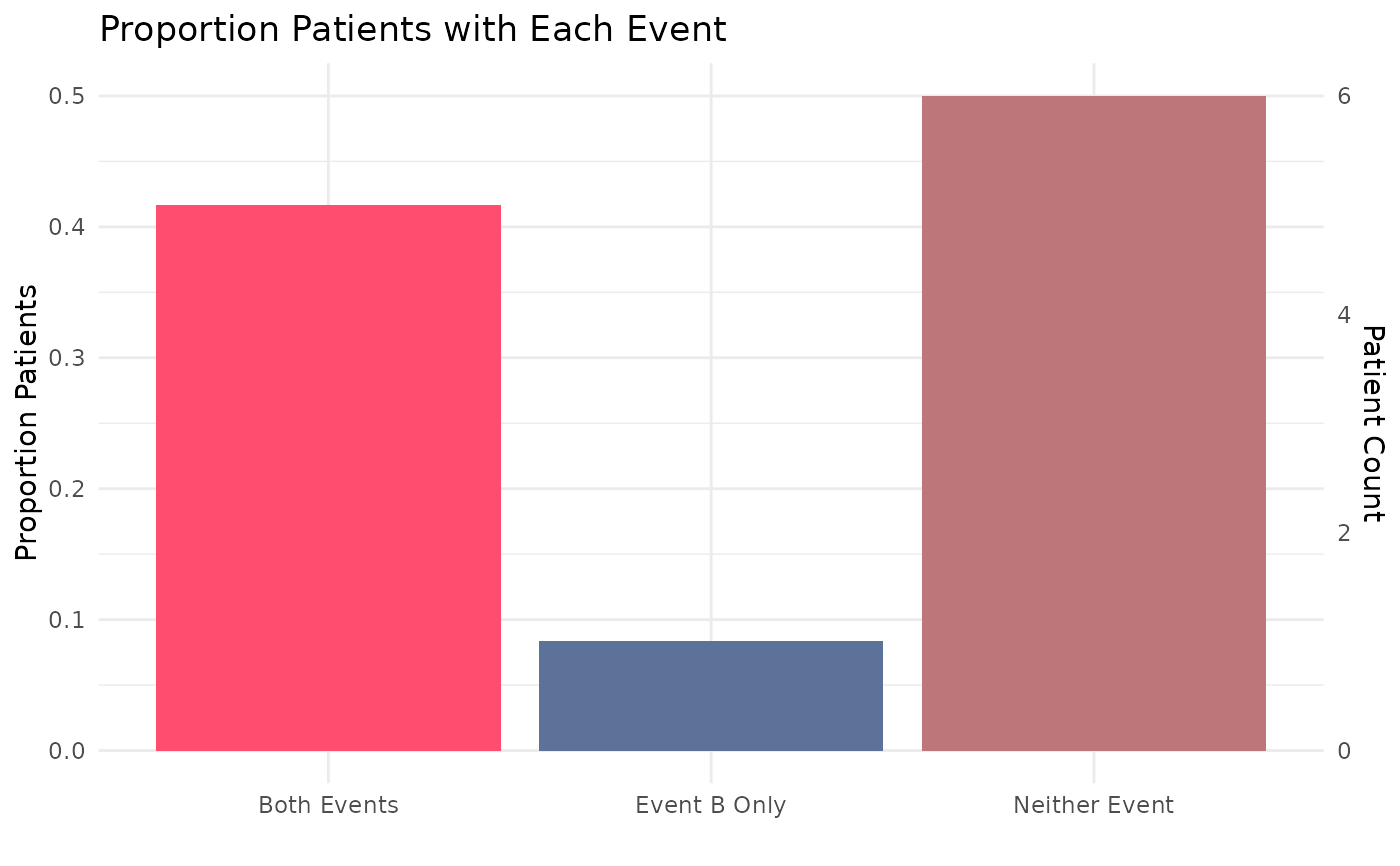 #' Easily convert the graph into an interactive ggiraph or plotly object with
#' `make_interactive_squba()`
make_interactive_squba(prc_output_example)
#' Easily convert the graph into an interactive ggiraph or plotly object with
#' `make_interactive_squba()`
make_interactive_squba(prc_output_example)